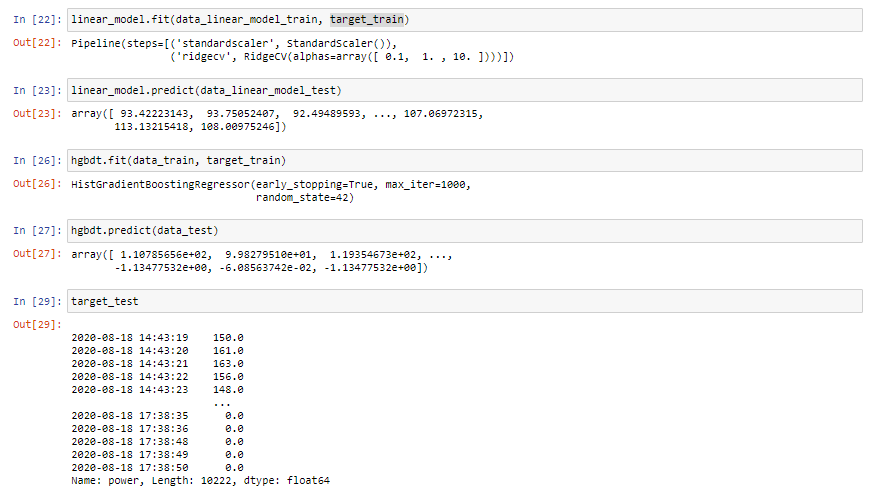
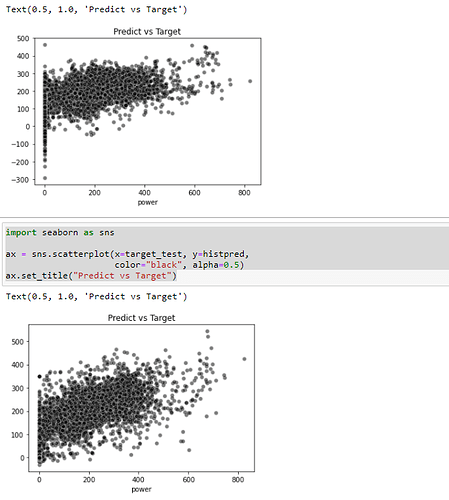
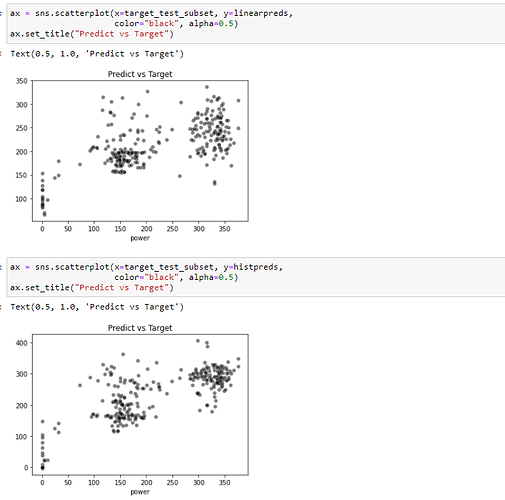
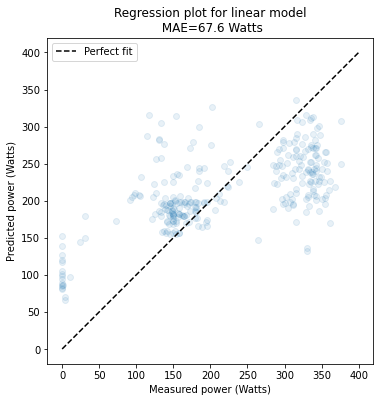
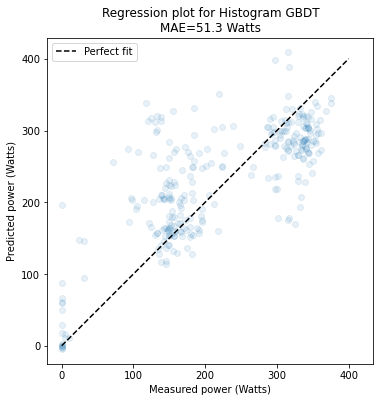
Excuse me, I have tried to fit the model with the code:
cv = LeaveOneGroupOut()
train_indices, test_indices = list(cv.split(data, target, groups=groups))[0]
data_linear_model_train = data_linear_model.iloc[train_indices]
data_linear_model_test = data_linear_model.iloc[test_indices]
data_train = data.iloc[train_indices]
data_test = data.iloc[test_indices]
target_train = target.iloc[train_indices]
target_test = target.iloc[test_indices]
cv_results_linear_model = cross_validate(
linear_model, data_linear_model_train, target_train, groups=groups, cv=cv,
scoring="neg_mean_absolute_error", return_estimator=True,
return_train_score=True, n_jobs=2)
cv_results_hgbdt = cross_validate(
hgbdt, data_train, target_train, groups=groups, cv=cv,
scoring="neg_mean_absolute_error", return_estimator=True,
return_train_score=True, n_jobs=2)
but I obtained:
ValueError: Found input variables with inconsistent numbers of samples: [28032, 28032, 38254]
It is possible I have omited some information?
Thanks