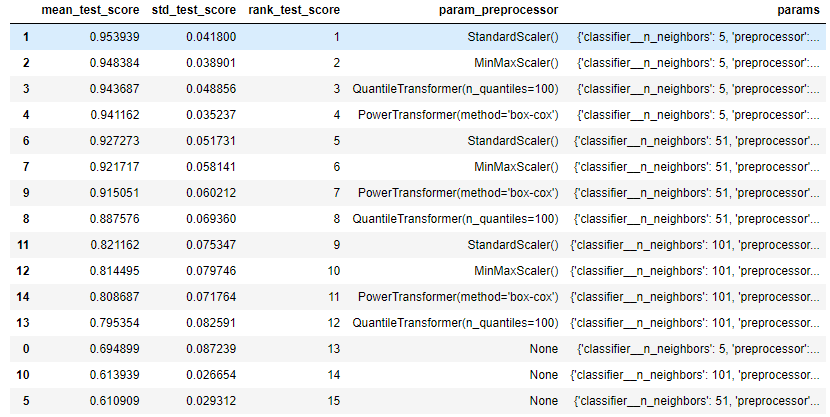
This is my code for Question 3. The question is asking us to evaluate the mean_test_score for different set of parameter from option a - b
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import QuantileTransformer
from sklearn.preprocessing import PowerTransformer
all_preprocessors = [
None,
StandardScaler(),
MinMaxScaler(),
QuantileTransformer(n_quantiles=100),
PowerTransformer(method="box-cox"),
]
from sklearn.model_selection import cross_validate
from sklearn.model_selection import GridSearchCV
from sklearn.neighbors import KNeighborsClassifier
from sklearn.pipeline import make_pipeline
model5 = make_pipeline(KNeighborsClassifier())
param_grid ={'preprocessor': all_preprocessors, 'classifier__n_neighbors': (5, 51, 101)}
model5_grid_search = GridSearchCV(model5, param_grid=param_grid, scoring='balanced_accuracy', cv=10)
There is an error when I run the following code:
model5_grid_search.fit(data, target)
The error is (it is a bit long so I just paste the ErrorValue statement):
ValueError: Invalid parameter classifier for estimator Pipeline(steps=[('kneighborsclassifier', KNeighborsClassifier())]). Check the list of available parameters with `estimator.get_params().keys()`.`
May I know what is my error about?
Secondly, when do we need to evaluate the model performance (particularly classification problem) using test_score, mean_test_score and standard_test_score?