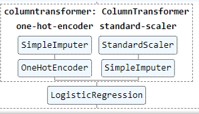
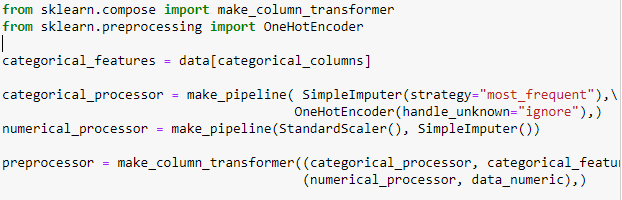
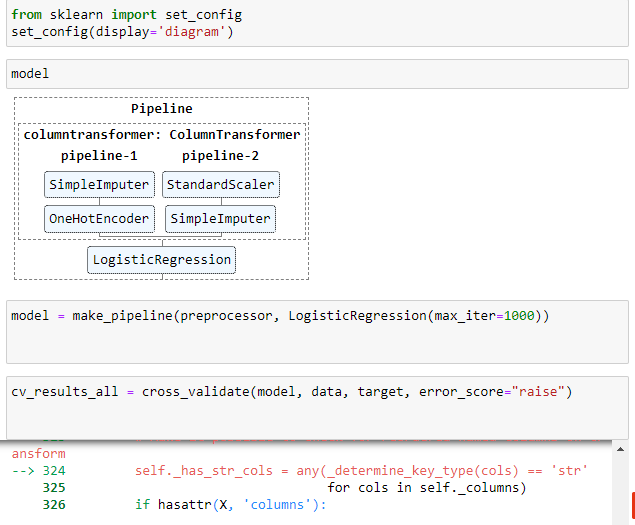
you are super fast 
The mean cross-validation accuracy is: nan +/- nan
/opt/conda/lib/python3.9/site-packages/sklearn/model_selection/_validation.py:615: FitFailedWarning: Estimator fit failed. The score on this train-test partition for these parameters will be set to nan. Details:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/sklearn/model_selection/_validation.py", line 598, in _fit_and_score
estimator.fit(X_train, y_train, **fit_params)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 341, in fit
Xt = self._fit(X, y, **fit_params_steps)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 303, in _fit
X, fitted_transformer = fit_transform_one_cached(
File "/opt/conda/lib/python3.9/site-packages/joblib/memory.py", line 352, in __call__
return self.func(*args, **kwargs)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 754, in _fit_transform_one
res = transformer.fit_transform(X, y, **fit_params)
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 505, in fit_transform
self._validate_remainder(X)
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 324, in _validate_remainder
self._has_str_cols = any(_determine_key_type(cols) == 'str'
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 324, in <genexpr>
self._has_str_cols = any(_determine_key_type(cols) == 'str'
File "/opt/conda/lib/python3.9/site-packages/sklearn/utils/__init__.py", line 268, in _determine_key_type
raise ValueError(err_msg)
ValueError: No valid specification of the columns. Only a scalar, list or slice of all integers or all strings, or boolean mask is allowed
warnings.warn("Estimator fit failed. The score on this train-test"
/opt/conda/lib/python3.9/site-packages/sklearn/model_selection/_validation.py:615: FitFailedWarning: Estimator fit failed. The score on this train-test partition for these parameters will be set to nan. Details:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/sklearn/model_selection/_validation.py", line 598, in _fit_and_score
estimator.fit(X_train, y_train, **fit_params)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 341, in fit
Xt = self._fit(X, y, **fit_params_steps)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 303, in _fit
X, fitted_transformer = fit_transform_one_cached(
File "/opt/conda/lib/python3.9/site-packages/joblib/memory.py", line 352, in __call__
return self.func(*args, **kwargs)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 754, in _fit_transform_one
res = transformer.fit_transform(X, y, **fit_params)
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 505, in fit_transform
self._validate_remainder(X)
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 324, in _validate_remainder
self._has_str_cols = any(_determine_key_type(cols) == 'str'
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 324, in <genexpr>
self._has_str_cols = any(_determine_key_type(cols) == 'str'
File "/opt/conda/lib/python3.9/site-packages/sklearn/utils/__init__.py", line 268, in _determine_key_type
raise ValueError(err_msg)
ValueError: No valid specification of the columns. Only a scalar, list or slice of all integers or all strings, or boolean mask is allowed
warnings.warn("Estimator fit failed. The score on this train-test"
/opt/conda/lib/python3.9/site-packages/sklearn/model_selection/_validation.py:615: FitFailedWarning: Estimator fit failed. The score on this train-test partition for these parameters will be set to nan. Details:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/sklearn/model_selection/_validation.py", line 598, in _fit_and_score
estimator.fit(X_train, y_train, **fit_params)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 341, in fit
Xt = self._fit(X, y, **fit_params_steps)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 303, in _fit
X, fitted_transformer = fit_transform_one_cached(
File "/opt/conda/lib/python3.9/site-packages/joblib/memory.py", line 352, in __call__
return self.func(*args, **kwargs)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 754, in _fit_transform_one
res = transformer.fit_transform(X, y, **fit_params)
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 505, in fit_transform
self._validate_remainder(X)
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 324, in _validate_remainder
self._has_str_cols = any(_determine_key_type(cols) == 'str'
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 324, in <genexpr>
self._has_str_cols = any(_determine_key_type(cols) == 'str'
File "/opt/conda/lib/python3.9/site-packages/sklearn/utils/__init__.py", line 268, in _determine_key_type
raise ValueError(err_msg)
ValueError: No valid specification of the columns. Only a scalar, list or slice of all integers or all strings, or boolean mask is allowed
warnings.warn("Estimator fit failed. The score on this train-test"
/opt/conda/lib/python3.9/site-packages/sklearn/model_selection/_validation.py:615: FitFailedWarning: Estimator fit failed. The score on this train-test partition for these parameters will be set to nan. Details:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/sklearn/model_selection/_validation.py", line 598, in _fit_and_score
estimator.fit(X_train, y_train, **fit_params)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 341, in fit
Xt = self._fit(X, y, **fit_params_steps)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 303, in _fit
X, fitted_transformer = fit_transform_one_cached(
File "/opt/conda/lib/python3.9/site-packages/joblib/memory.py", line 352, in __call__
return self.func(*args, **kwargs)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 754, in _fit_transform_one
res = transformer.fit_transform(X, y, **fit_params)
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 505, in fit_transform
self._validate_remainder(X)
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 324, in _validate_remainder
self._has_str_cols = any(_determine_key_type(cols) == 'str'
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 324, in <genexpr>
self._has_str_cols = any(_determine_key_type(cols) == 'str'
File "/opt/conda/lib/python3.9/site-packages/sklearn/utils/__init__.py", line 268, in _determine_key_type
raise ValueError(err_msg)
ValueError: No valid specification of the columns. Only a scalar, list or slice of all integers or all strings, or boolean mask is allowed
warnings.warn("Estimator fit failed. The score on this train-test"
/opt/conda/lib/python3.9/site-packages/sklearn/model_selection/_validation.py:615: FitFailedWarning: Estimator fit failed. The score on this train-test partition for these parameters will be set to nan. Details:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/sklearn/model_selection/_validation.py", line 598, in _fit_and_score
estimator.fit(X_train, y_train, **fit_params)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 341, in fit
Xt = self._fit(X, y, **fit_params_steps)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 303, in _fit
X, fitted_transformer = fit_transform_one_cached(
File "/opt/conda/lib/python3.9/site-packages/joblib/memory.py", line 352, in __call__
return self.func(*args, **kwargs)
File "/opt/conda/lib/python3.9/site-packages/sklearn/pipeline.py", line 754, in _fit_transform_one
res = transformer.fit_transform(X, y, **fit_params)
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 505, in fit_transform
self._validate_remainder(X)
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 324, in _validate_remainder
self._has_str_cols = any(_determine_key_type(cols) == 'str'
File "/opt/conda/lib/python3.9/site-packages/sklearn/compose/_column_transformer.py", line 324, in <genexpr>
self._has_str_cols = any(_determine_key_type(cols) == 'str'
File "/opt/conda/lib/python3.9/site-packages/sklearn/utils/__init__.py", line 268, in _determine_key_type
raise ValueError(err_msg)
ValueError: No valid specification of the columns. Only a scalar, list or slice of all integers or all strings, or boolean mask is allowed
warnings.warn("Estimator fit failed. The score on this train-test"
```