I have tried to use the KBinsDescretizer and the Nystroem preprocessors to make a linear model of piecewise polynomials of a given degree (typically one or two) but it did not work, I only got the same results. How could I create such a model ?
Let’s try a toy example. We generate the following data:
import numpy as np
x = np.linspace(0, 10, num=500)
X = x.reshape(-1, 1).copy()
y = []
for xx in x:
if xx < 3:
y.append(xx ** 3)
elif xx > 3 and xx < 7:
y.append(xx)
else:
y.append(xx ** 2)
You could define the pipeline in this manner:
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import KBinsDiscretizer, PolynomialFeatures
from sklearn.kernel_approximation import Nystroem
from sklearn.linear_model import LinearRegression
model = make_pipeline(
Nystroem(kernel="poly", degree=3),
KBinsDiscretizer(n_bins=3, strategy="uniform"),
LinearRegression(),
).fit(X, y)
y_pred = model.predict(X)
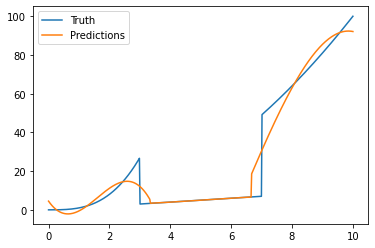
We plot the results:
import matplotlib.pyplot as plt
plt.plot(x, y, label="Truth")
plt.plot(x, y_pred, label="Predictions")
_ = plt.legend()
Thank you Guillaume.
This is not quite what I wanted, because the solution you provide still find a piecewise constant solution. What I was trying to do is to find boundaries between the domains (here x_1=3 and x_2=7) and the coefficients a_i, b_i, c_i and d_i with i = 1,2,3 such that
y = a_i x³ + b_i x² + c_i x + d_i in the range I_i
on each of the three ranges I_1=[0, x_1], I_2=[x_1, x_2] and I_3=[x_2, 10].
It seems that the purpose of the ‘poly’ kernel in Nystroem is to combine the coordinates in polynomials to divide space into domains with non-straight boundaries.
Nystroem is a kernel approximation. A kernel is usually defined as K(x, x'). Therefore, this is a product between data-point. I am under the impression that you want instead a PolynominalFeatures.
The issue there is that KBinsDiscretizer is one hot encoded. Therefore, you get 3 columns for each interval in X but the values are either 0 or 1. Applying a polynomial will still result in 0 or 1 value per column. Therefore, you can only predict a piecewise constant.
So if you want to make a piecewise polynomial, you need to modify the KBinsDiscretizer such that the encoding is multiplied by the original data. In this case, you need your own transformer because scikit-learn does not provide one. Here, is a really unoptimized version of it:
from scipy import sparse
from sklearn.preprocessing import KBinsDiscretizer
class MyTransformer(KBinsDiscretizer):
def fit(self, X, y):
if self.encode not in ("onehot", "onehot-dense"):
raise ValueError(
"This transformer only works with one hot encoded discretization"
)
return super().fit(X, y)
def transform(self, X):
X_trans = super().transform(X)
n_bins_cumsum = np.insert(np.cumsum(self.n_bins_), 0, 0)
for idx in range(n_bins_cumsum.size - 1):
if sparse.issparse(X_trans):
X_trans[:, n_bins_cumsum[idx]:n_bins_cumsum[idx + 1]] = (
X_trans[:, n_bins_cumsum[idx]:n_bins_cumsum[idx + 1]].multiply(X[:, idx].reshape(-1, 1))
)
else:
X_trans[:, n_bins_cumsum[idx]:n_bins_cumsum[idx + 1]] *= X[:, idx].reshape(-1, 1)
return X_trans
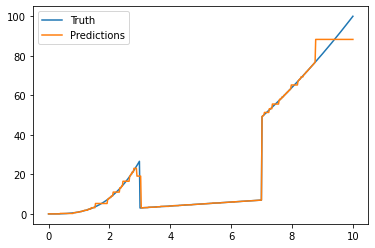
Then one can construct a new pipeline:
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import KBinsDiscretizer, PolynomialFeatures
from sklearn.linear_model import LinearRegression
model = make_pipeline(
MyTransformer(n_bins=3, strategy="uniform"),
PolynomialFeatures(degree=3),
LinearRegression(),
).fit(X, y)
y_pred = model.predict(X)
and get the following plot: